 |
||
|
|
||
|
Page Title:
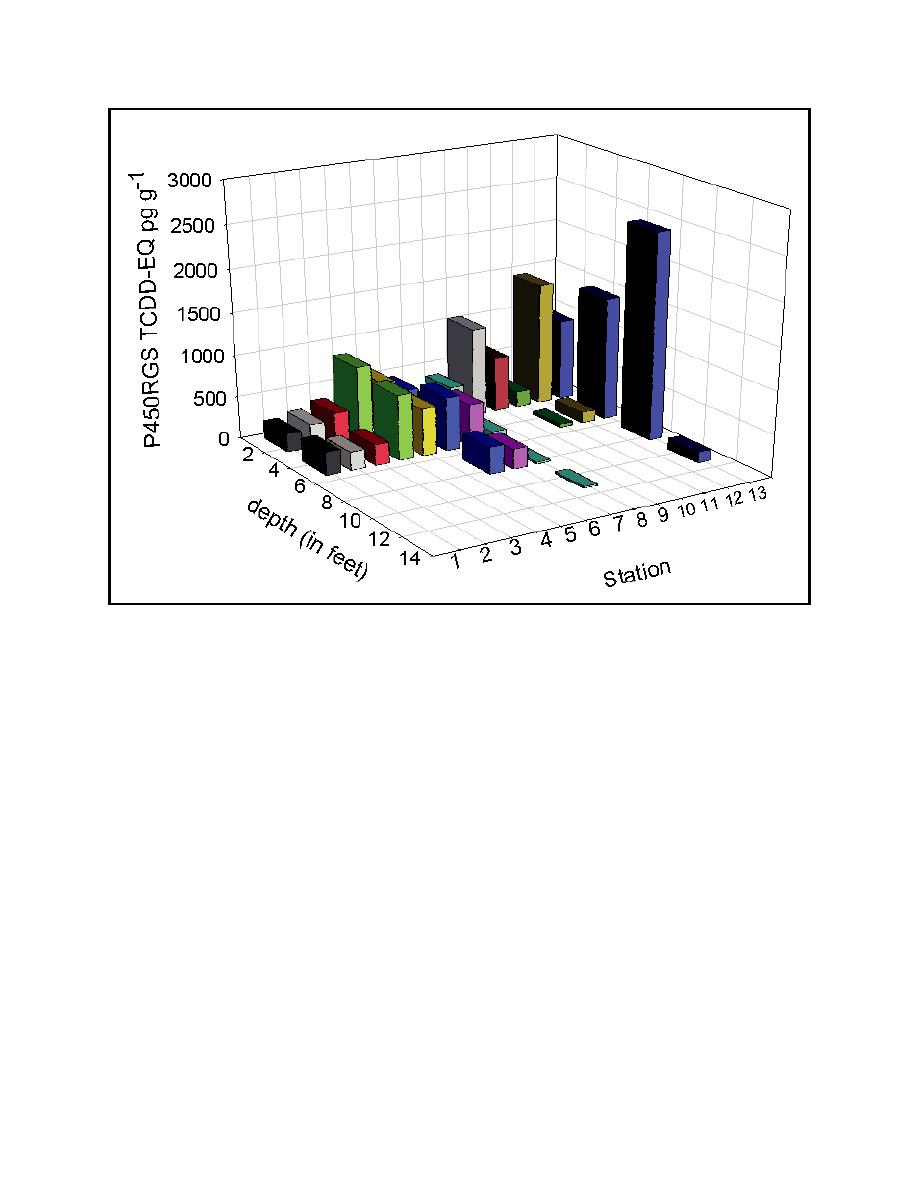
Figure 3. The depth profile of TCDD-EQ at each site as determined by the P450RGS assay. |
||
| |||||||||||||||
|
|
 ERDC TN-DOER-C19
December 2000
Figure 3. The depth profile of TCDD-EQ at each site as determined by the P450RGS assay. Each bar
represents an average from three extracts from each of the five cores (15 samples per depth
per site). Note that at several stations the contamination varies with depth; such information
could not be obtained from chemical analysis alone due to prohibitively high costs
the detail obtained with this cell-based assay would have been prohibitively expensive using
GC-MS, costing at least $558,000 (155 samples with triplicate extractions, $1,200 per analysis)
compared with the $31,000 spent on the cell-based assay ($200 per sample, includes analysis of trip-
licate extractions).
Cell-based assays such as P450RGS can also be used to determine the bioavailability of contami-
nants, by analyzing both the media (e.g., water, soil, sediment) and organism (e.g., fish, worm, or
clam) of interest. Several currently proposed projects plan on using the cell-based assay to monitor
the efficacy of remediation techniques for polycyclic aromatic hydrocarbons (PAHs),
polychlorinated biphenyls (PCBs), and dioxins by exposing organisms to the soil or sediment be-
fore, during, and after remediation, then extracting them and analyzing the extracts with the
cell-based assay.
Development of New Cell-Based Assays. Cell-based techniques similar to that of the
P450RGS assay can also be developed for other contaminant classes. The metal-responsive
metallothionein gene is characterized well enough that a cell-based assay can be developed in a man-
ner similar to the P450RGS system (Iyengar, Muller, and Maclean 1996). Another class of
4
|
|
Privacy Statement - Press Release - Copyright Information. - Contact Us - Support Integrated Publishing |