 |
||
|
|
||
|
Page Title:
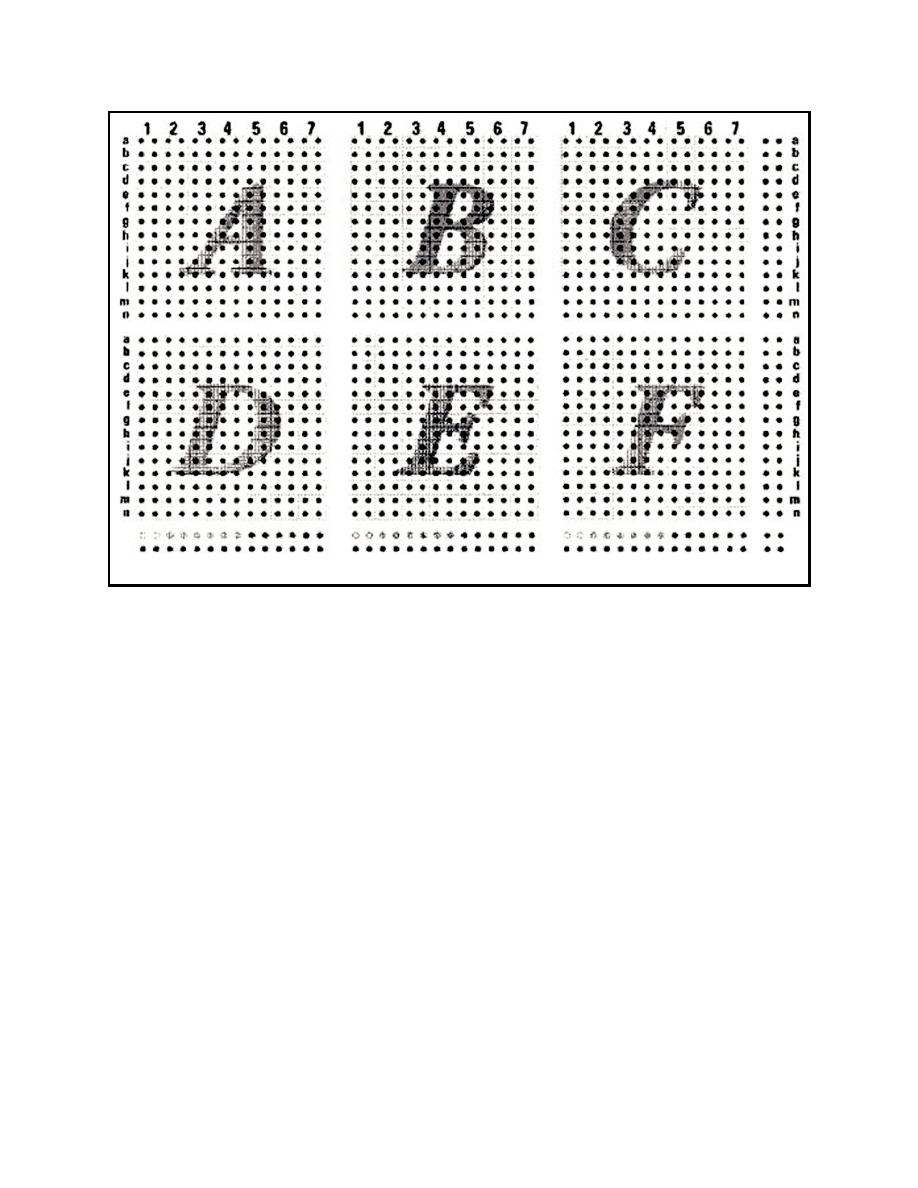
Figure 5. An example of a cDNA array: the Clontech Human Toxicology Array II. |
||
| |||||||||||||||
|
|
 ERDC TN-DOER-C19
December 2000
Figure 5. An example of a cDNA array: the Clontech Human Toxicology Array II. Each spot represents
a selected toxicologically related gene bound to a nylon membrane. Genes in area A are
related to oncogenes, tumor supressors, cell cycle regulators, and ion channels; area B
contains genes related to transporters and signal transduction; area C contains genes related
to apoptosis, DNA synthesis, repair, and recombination; area D contains genes related to the
nervous system and transcription factors; area E contains genes related to various receptors
(e.g., growth factors, hormones), cell adhesion, and general DNA binding proteins; area F
contains genes related to stress response, cell signaling/cell-cell communication, and protein
turnover
Current Status/Future Directions. At this point, only preliminary trials utilizing the Clontech
membrane arrays have been conducted, with initial attempts made to develop a chemiluminescent
endpoint rather than the standard radiometric endpoint recommended by the company. Although
the results were not promising (high background, low sensitivity), Clontech has recently released a
glass array for which chemiluminescent and fluorescent endpoints have been developed. These new
developments will be used in conjunction with the hepG2 cell line to develop profiles of genes re-
sponding to various common pollutants encountered in dredged materials, as well as the magnitude
of the response in relation to the dose. This information, along with the response of the array to
dredged material extracts, can eventually be utilized to tailor an array with specific genes of interest
that provide data that best fulfill the U.S. Army Corps of Engineers risk assessment needs.
CONCLUSIONS: By analyzing the response of cells to extracts of sediment and/or tissue, cDNA
arrays will provide a far better measure of exposure and effect for risk characterization and input to
8
|
|
Privacy Statement - Press Release - Copyright Information. - Contact Us - Support Integrated Publishing |