 |
||
|
|
||
|
Page Title:
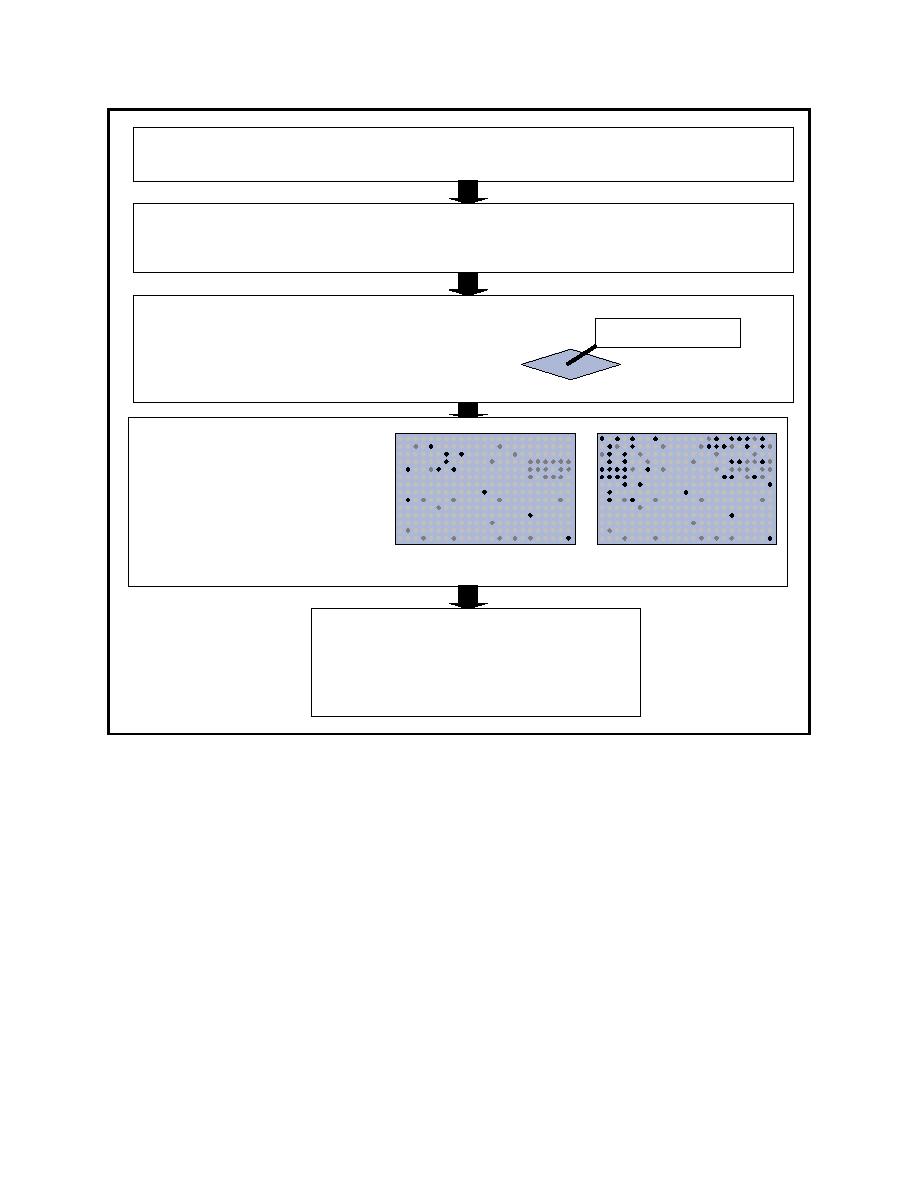
Figure 6. An overview of the cDNA array assay. |
||
| |||||||||||||||
|
|
 ERDC TN-DOER-C19
December 2000
Isolate mRNA population
AAAUGUCGGA...
Convert to labeled cDNA
AAAUGUCGGA...
(PCR)
TTTACAGCCT...
Hybridize to array
TTTACAGCCT...
AAATGTCGGA...
Array membrane
Detection of labeled cDNA
(radiometric, chemiluminescent,
or fluorescent detection)
Treated
Control
Compare control and treated arrays to
determine which genes have been up or
down regulated (increase or decreased
signal compared to control array)
Figure 6. An overview of the cDNA array assay. The mRNA populations are isolated from control and
treated cell cultures (a partial sequence shown to the right as an example), converted to
tagged cDNA (shown in green), and hybridized to the cDNA bound to the array (one of the
genes on the array shown in blue)
models for risk assessment than is presently possible (Corton et al. 1999; Nuwaysir et al. 1999;
Rockett and Dix 1999). The mechanistic information obtained from the arrays can also be valuable
in determining which samples require further examination with more time-consuming and expen-
sive bioassays, and can direct the type of bioassay (e.g., survival, growth and reproduction,
genotoxicity) that should be conducted. Additionally, use of the cDNA arrays to test sediment ex-
tracts can help direct the development and/or use of other assays. For example, screening of typical
dredged sediments with cDNA arrays may indicate that certain endpoints (e.g., metal-responsive
genes) consistently show positive responses and warrant further evaluation with cell-based assays,
which can provide high throughput screens due to their compatibility with the 96-well microtiter
plate format. Alternatively, initial screening of a site with cDNA arrays may indicate that a certain
contaminant class is of concern, whereupon the appropriate cell-based assays can be utilized to
9
|
|
Privacy Statement - Press Release - Copyright Information. - Contact Us - Support Integrated Publishing |